Frankenstein’s…bacteriophages?

Frankenstein’s…bacteriophages?
A group of scientists have just published their work in CellPress on synthesising novel phages using common viral structures (Figure 1), to target a range of bacterial hosts. Anyone who frequently reads my posts will be fully aware of what bacteriophages are, however put simply phages are viruses that target bacteria (Figure 1). Currently a lot of research is going into these little guys to utilise them for both therapeutic and diagnostic purposes.

Figure 1. Image depicts a transmission electron micrograph of a phage (left) and a diagrammatic representation of the phage on the (right) showing all the major components that make up the protein coat.
Phages have an elaborate protein coat that surrounds their DNA cargo (contained within the head, Figure 1). They scan their surrounding environment for potential ‘prey’ by using their tail fibres to interact with their cognate receptors. The diversity of receptors is immense and many still go unidentified even for well characterised phages. Once a target is found, the baseplate then irreversibly attaches to the cell wall of the bacteria and the DNA can be transferred into the cytoplasm of the cell.
Unlike current therapeutics, the sheer diversity between phages means that they are highly specific with limited host ranges to the species or even strain level. As a result, for phage therapy to be effective, a cocktail of phages needs to be employed to target a wide range of potential bacterial hosts. To improve the host ranges of phage groups such as this one are trying to synthesis novel structures from existing phages - much like how Frankenstein stitched together a new human body from pre-existing parts (Figure 2).

Figure 2. Simplified image showing how bacterial components can be shuffled between genomes of related phages with different host ranges.
Innovatively, this group used a well characterised yeast-based (Saccharomyces cerevisiae) platform for capturing phage genomes to allow their genetic manipulation (Figure 3). Phage genomes could then be inserted into a yeast artificial chromosome (YAC) then manipulated. Yeast are fairly easy to genetically modify by homologous-recombingation, making this system far easier to employ than other methods. The YAC was then recovered and transformed into the normal host bacteria, allowing the generation of new phage particles. Unlike other methods phage generated via this method were relatively easy to reboot back into active viral particles.

Figure 3. Illustration of the genetic manipulation of phage DNA to generate hybrid phages with altered host ranges.
The group managed to show that by swapping modular components such as tail fibres between phages, new host specificity could be generated. Thus illustrating that gene swapping can overcome strain or species barriers if the need arises. This work will hopefully lead way to further improve phage therapy and decrease the persistent need for the identification of novel phages. Further work needs to be done on increasing the scope of their work, but they have created a framework that will hopefully be able to reboot more synthetic phages.
“Our results show that common phage scaffolds can be re-targeted against new bacterial hosts by engineering single or multiple tail components. This capability enables the the construction of defined phage cocktails that only differ in their host range determinants and can be used to edit the compositions of microbial consortia and/or treat bacterial infections.” - Ando et al, 2015
Sources:
Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing, 2015. Ando, H., Lemire. S., Pires. D.P., Lu. T.K., Cell Systems , Volume 1 , Issue 3 , 187 - 196
More Posts from Fuadalanazi and Others


Bacterias & Tx
Hi guys, just wanted to let you know that I did this really fast, so if there’s something wrong let me know! thanks :)
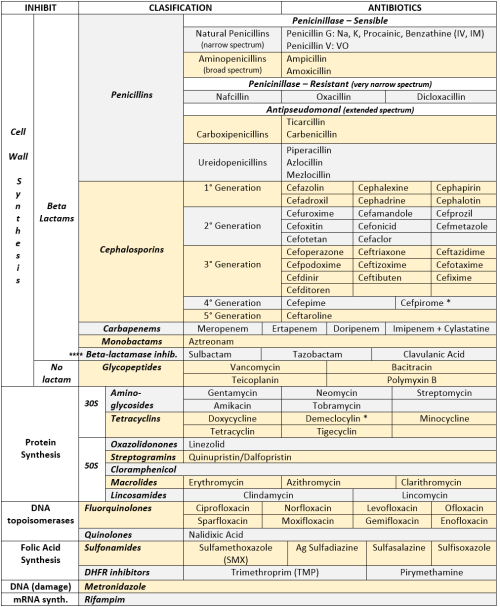
ANTIBIOTICS CHEAT SHEET :)
Also, REMEMBER!!!!
* Sulfonamides compete for albumin with:
Bilirrubin: given in 2°,3°T, high risk or indirect hyperBb and kernicterus in premies
Warfarin: increases toxicity: bleeding
* Beta-lactamase (penicinillase) Suceptible:
Natural Penicillins (G, V, F, K)
Aminopenicillins (Amoxicillin, Ampicillin)
Antipseudomonal Penicillins (Ticarcillin, Piperacillin)
* Beta-lactamase (penicinillase) Resistant:
Oxacillin, Nafcillin, Dicloxacillin
3°G, 4°G Cephalosporins
Carbapenems
Monobactams
Beta-lactamase inhibitors
* Penicillins enhanced with:
Clavulanic acid & Sulbactam (both are suicide inhibitors, they inhibit beta-lactamase)
Aminoglycosides (against enterococcus and psedomonas)
* Aminoglycosides enhanced with Aztreonam
* Penicillins: renal clearance EXCEPT Oxacillin & Nafcillin (bile)
* Cephalosporines: renal clearance EXCEPT Cefoperazone & Cefrtriaxone (bile)
* Both inhibited by Probenecid during tubular secretion.
* 2°G Cephalosporines: none cross BBB except Cefuroxime
* 3°G Cephalosporines: all cross BBB except Cefoperazone bc is highly highly lipid soluble, so is protein bound in plasma, therefore it doesn’t cross BBB.
* Cephalosporines are "LAME“ bc they do not cover this organisms
L isteria monocytogenes
A typicals (Mycoplasma, Chlamydia)
M RSA (except Ceftaroline, 5°G)
E nterococci

* Disulfiram-like effect: Cefotetan & Cefoperazone (mnemonic)
* Cefoperanzone: all the exceptions!!!
All 3°G cephalosporins cross the BBB except Cefoperazone.
All cephalosporins are renal cleared, except Cefoperazone.
Disulfiram-like effect
* Against Pseudomonas:
3°G Cef taz idime (taz taz taz taz)
4°G Cefepime, Cefpirome (not available in the USA)
Antipseudomonal penicillins
Aminoglycosides (synergy with beta-lactams)
Aztreonam (pseudomonal sepsis)
* Covers MRSA: Ceftaroline (rhymes w/ Caroline, Caroline the 5°G Ceph), Vancomycin, Daptomycin, Linezolid, Tigecycline.
* Covers VRSA: Linezolid, Dalfopristin/Quinupristin
* Aminoglycosides: decrease release of ACh in synapse and act as a Neuromuscular blocker, this is why it enhances effects of muscle relaxants.
* DEMECLOCYCLINE: tetracycline that’s not used as an AB, it is used as tx of SIADH to cause Nephrogenic Diabetes Insipidus (inhibits the V2 receptor in collecting ducts)
* Phototoxicity: Q ue S T ion?
Q uinolones
Sulfonamides
T etracyclines

* p450 inhibitors: Cloramphenicol, Macrolides (except Azithromycin), Sulfonamides
* Macrolides SE: Motilin stimulation, QT prolongation, reversible deafness, eosinophilia, cholestatic hepatitis
* Bactericidal: beta-lactams (penicillins, cephalosporins, monobactams, carbapenems), aminoglycosides, fluorquinolones, metronidazole.
* Baceriostatic: tetracyclins, streptogramins, chloramphenicol, lincosamides, oxazolidonones, macrolides, sulfonamides, DHFR inhibitors.
* Pseudomembranous colitis: Ampicillin, Amoxicillin, Clindamycin, Lincomycin.
* QT prolongation: macrolides, sometimes fluoroquinolones

يالله 💙

JUST THE FAQS: Top Scientists Around the Nation Call for a Unified Microbiome Initiative
Writing in the journal Science this week, a group of top microbiome researchers from around the country, including UC San Diego School of Medicine’s Rob Knight, PhD, and Pieter Dorrestein, PhD, lay out a proposal for what they call a Unified Microbiome Initiative. This latest post in the JUST THE FAQS series takes a look at what that means and why it’s important.
What is the Unified Microbiome Initiative?
The proposed Unified Microbiome Initiative is a national effort to fund, coordinate and accelerate microbiome research — the study of the microbial communities (bacteria, viruses, etc.) that live in, on and around us.
Why do we need this?
According to the authors of this proposal, the microbiome field currently lacks many of the technologies and resources required to take the research from where it is today (i.e., mostly describing what microbes are living where) to the next level — understanding exactly how microbes influence our health and how we might be able to manipulate them to our benefit.
“… such knowledge could transform our understanding of the world and launch innovations in agriculture, energy, health, the environment and more,” the authors write in the proposal.
This ambitious undertaking cannot be accomplished by individual laboratories working in isolation. Developing these tools requires new collaborations between physical, life, and biomedical sciences, engineering and many other disciplines. Advancing this nascent field also depends on attracting, training and supporting multidisciplinary networks of scientists and engineers, all things that could be accomplished through a concerted national effort, Knight, Dorrestein and their co-authors say.
Wait, what is the microbiome again?
Your body contains as many as 10 times more microbial cells than human cells. Even crazier, that adds up to two to 20 million microbial genes, compared to your 20,000 or so human genes. By that measure, you could say you’re actually only about 10 percent human.
Researchers like Knight, Dorrestein and their teams are now mapping the other 90 percent — the collections of microbial genes (“microbiomes”) found in our guts, mouths, skin, and many other locations on and around the body. They’re finding that the makeup of our gut microbiomes is associated with a rapidly growing list of diseases and conditions: food allergies, obesity, inflammatory bowel disease and colon cancer, rheumatoid arthritis, atherosclerosis, asthma, perhaps even anxiety, depression and autism. But researchers don’t yet know why these associations occur, whether they are cause or effect, and whether or not we could use gut microbiome readouts for medical predictions, diagnoses or treatments.
And that’s just the gut microbiome. Imagine what researchers will find when they dive as deeply into the microbiomes of the rest of our bodies, as well as our homes, the ocean and other environments. That’s why Knight, Dorrestein and many other experts are today calling for a collaborative, well-supported United Microbiome Initiative.
To learn more about Knight’s and Dorrestein’s microbiome work, check out: Antibiotic Resistance in Microbiomes, Despite Isolation from Western Civilization
3D Human Skin Maps Aid Study of Relationships Between Molecules, Microbes and Environment
Pictured: Staphylococcus aureus bacteria. Source: NIAID

“Sequence VR" was exhibited at the V & A Museum in London, as part of the London Design Festival. A new immersive virtual reality experience has been created using an Oculus Rift as part of “Sequence” using the data and footage from the project. It was shown alongside a series of objects and artefacts created during the project including live bacteria. The exhibit was accompanied by a participatory DNA extraction/preparation workshop where Dumitriu was joined by Dr Nicola Fawcett from the Modernisng Medical Microbiology Project. The event took place on 25th - 27th September 2015. See more information here.
http://www.youtube.com/embed/TS0zA6DSfvk?feature=oembed
الرقية الشرعية الشاملة فارس عباد 1 (by netter2cool)
Penicillin

Penicillin is a widely used antibiotic prescribed to treat staphylococci and streptococci bacterial infections.
beta-lactam family
Gram-positive bacteria = thick cell walls containing high levels of peptidoglycan
gram-negative bacteria = thinner cell walls with low levels of peptidoglycan and surrounded by a lipopolysaccharide (LPS) layer that prevents antibiotic entry
penicillin is most effective against gram-positive bacteria where DD-transpeptidase activity is highest.
Examples of penicillins include:
amoxicillin
ampicillin
bacampicillin
oxacillin
penicillin
Mechanism(s)
Penicillin inhibits the bacterial enzyme transpeptidase, responsible for catalysing the final peptidoglycan crosslinking stage of bacterial cell wall synthesis.
Cells wall is weakened and cells swell as water enters and then burst (lysis)
Becomes permanently covalently bonded to the enzymes’s active site (irreversible)


Alternative theory: penicillin mimics D-Ala D-Ala

Or may act as an umbrella inhibitor

Resistance
production of beta-lactamase - destroys the beta-lactam ring of penicillin and makes it ineffective (eg Staphylococcus aureus - most are now resistant)
In response, synthetic penicillin that is resistant to beta-lactamase is in use including egdicloxacillin, oxacillin, nafcillin, and methicillin.
Some is resistant to methicillin - methicillin-resistant Staphylococcus aureus (MRSA).
Demonstrating blanket resistance to all beta-lactam antibiotics -extremely serious health risk.
-
 perpetualstudentzero reblogged this · 8 years ago
perpetualstudentzero reblogged this · 8 years ago -
 perpetualstudentzero reblogged this · 8 years ago
perpetualstudentzero reblogged this · 8 years ago -
 perpetualstudentzero reblogged this · 8 years ago
perpetualstudentzero reblogged this · 8 years ago -
 chicagowindchill reblogged this · 9 years ago
chicagowindchill reblogged this · 9 years ago -
 tayywiddd-blog liked this · 9 years ago
tayywiddd-blog liked this · 9 years ago -
 groaninggranada liked this · 9 years ago
groaninggranada liked this · 9 years ago -
 srgcpzz liked this · 9 years ago
srgcpzz liked this · 9 years ago -
 space-caramel liked this · 9 years ago
space-caramel liked this · 9 years ago -
 whododis liked this · 9 years ago
whododis liked this · 9 years ago -
 mrbillybd reblogged this · 9 years ago
mrbillybd reblogged this · 9 years ago -
 apathetic182 reblogged this · 9 years ago
apathetic182 reblogged this · 9 years ago -
 study-biotechnology-blog liked this · 9 years ago
study-biotechnology-blog liked this · 9 years ago -
 casuallyjollybird liked this · 9 years ago
casuallyjollybird liked this · 9 years ago -
 bookaboutflames liked this · 9 years ago
bookaboutflames liked this · 9 years ago -
 archiveofbiognosis reblogged this · 9 years ago
archiveofbiognosis reblogged this · 9 years ago -
 mcclintocks reblogged this · 9 years ago
mcclintocks reblogged this · 9 years ago -
 opdroygaraytekin reblogged this · 9 years ago
opdroygaraytekin reblogged this · 9 years ago -
 opdroygaraytekin liked this · 9 years ago
opdroygaraytekin liked this · 9 years ago -
 queenofeire liked this · 9 years ago
queenofeire liked this · 9 years ago -
 jerkymate liked this · 9 years ago
jerkymate liked this · 9 years ago -
 madamewigglymon reblogged this · 9 years ago
madamewigglymon reblogged this · 9 years ago -
 awh-george-blog reblogged this · 9 years ago
awh-george-blog reblogged this · 9 years ago -
 megres liked this · 9 years ago
megres liked this · 9 years ago -
 geiszler reblogged this · 9 years ago
geiszler reblogged this · 9 years ago -
 brokenergy liked this · 9 years ago
brokenergy liked this · 9 years ago -
 brokenergy reblogged this · 9 years ago
brokenergy reblogged this · 9 years ago -
 nezya liked this · 9 years ago
nezya liked this · 9 years ago -
 destroywhatdestroysus reblogged this · 9 years ago
destroywhatdestroysus reblogged this · 9 years ago -
 destroywhatdestroysus liked this · 9 years ago
destroywhatdestroysus liked this · 9 years ago -
 sininenvariksen reblogged this · 9 years ago
sininenvariksen reblogged this · 9 years ago -
 sininenvariksen liked this · 9 years ago
sininenvariksen liked this · 9 years ago -
 corasombrelune liked this · 9 years ago
corasombrelune liked this · 9 years ago -
 samsmithsam-blog liked this · 9 years ago
samsmithsam-blog liked this · 9 years ago -
 monirosez reblogged this · 9 years ago
monirosez reblogged this · 9 years ago -
 monirosez liked this · 9 years ago
monirosez liked this · 9 years ago -
 lasenbyphoenix liked this · 9 years ago
lasenbyphoenix liked this · 9 years ago -
 even-stumbling-is-progress reblogged this · 9 years ago
even-stumbling-is-progress reblogged this · 9 years ago -
 ruigglechrome liked this · 9 years ago
ruigglechrome liked this · 9 years ago -
 dlane247 liked this · 9 years ago
dlane247 liked this · 9 years ago