(via Https://www.youtube.com/watch?v=TkdZ8k4VHZ4)
(via https://www.youtube.com/watch?v=TkdZ8k4VHZ4)
More Posts from Fuadalanazi and Others
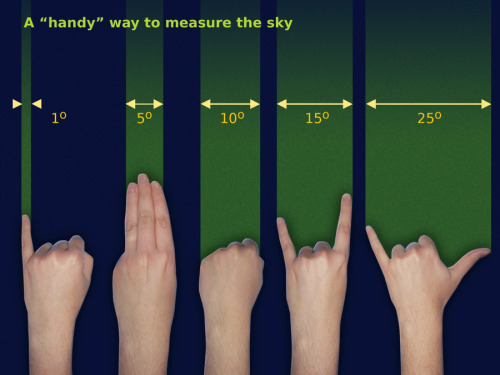
Measuring the sky, the handy way.
Source: Free Astronomy Teaching Resources (Starry Night)
A Basic Demonstration of Optical Cloaking (by UniversityRochester)

JUST THE FAQS: Top Scientists Around the Nation Call for a Unified Microbiome Initiative
Writing in the journal Science this week, a group of top microbiome researchers from around the country, including UC San Diego School of Medicine’s Rob Knight, PhD, and Pieter Dorrestein, PhD, lay out a proposal for what they call a Unified Microbiome Initiative. This latest post in the JUST THE FAQS series takes a look at what that means and why it’s important.
What is the Unified Microbiome Initiative?
The proposed Unified Microbiome Initiative is a national effort to fund, coordinate and accelerate microbiome research — the study of the microbial communities (bacteria, viruses, etc.) that live in, on and around us.
Why do we need this?
According to the authors of this proposal, the microbiome field currently lacks many of the technologies and resources required to take the research from where it is today (i.e., mostly describing what microbes are living where) to the next level — understanding exactly how microbes influence our health and how we might be able to manipulate them to our benefit.
“… such knowledge could transform our understanding of the world and launch innovations in agriculture, energy, health, the environment and more,” the authors write in the proposal.
This ambitious undertaking cannot be accomplished by individual laboratories working in isolation. Developing these tools requires new collaborations between physical, life, and biomedical sciences, engineering and many other disciplines. Advancing this nascent field also depends on attracting, training and supporting multidisciplinary networks of scientists and engineers, all things that could be accomplished through a concerted national effort, Knight, Dorrestein and their co-authors say.
Wait, what is the microbiome again?
Your body contains as many as 10 times more microbial cells than human cells. Even crazier, that adds up to two to 20 million microbial genes, compared to your 20,000 or so human genes. By that measure, you could say you’re actually only about 10 percent human.
Researchers like Knight, Dorrestein and their teams are now mapping the other 90 percent — the collections of microbial genes (“microbiomes”) found in our guts, mouths, skin, and many other locations on and around the body. They’re finding that the makeup of our gut microbiomes is associated with a rapidly growing list of diseases and conditions: food allergies, obesity, inflammatory bowel disease and colon cancer, rheumatoid arthritis, atherosclerosis, asthma, perhaps even anxiety, depression and autism. But researchers don’t yet know why these associations occur, whether they are cause or effect, and whether or not we could use gut microbiome readouts for medical predictions, diagnoses or treatments.
And that’s just the gut microbiome. Imagine what researchers will find when they dive as deeply into the microbiomes of the rest of our bodies, as well as our homes, the ocean and other environments. That’s why Knight, Dorrestein and many other experts are today calling for a collaborative, well-supported United Microbiome Initiative.
To learn more about Knight’s and Dorrestein’s microbiome work, check out: Antibiotic Resistance in Microbiomes, Despite Isolation from Western Civilization
3D Human Skin Maps Aid Study of Relationships Between Molecules, Microbes and Environment
Pictured: Staphylococcus aureus bacteria. Source: NIAID
BugsFeed: 7 bad ass organisms that can survive intracellularly in immune cells
1. Mycobacterium tuberculosis - Stops fusion!

Mycobacterium tuberculosis utilizes macrophages for its replication! (It uses the usual killer to expand it’s army :O ) How does tuberculosis bacilli survive in macrophages? M. tuberculosis has evolved a number of very effective survival strategies - It inhibits phagosome-lysosome fusion and inhibits phagosome acidification ensuring it’s survival inside the macrophage.
2. Brucella - Has chains, like Bruce Lee.

Brucella has a LPS O-chain. It ensures the Brucella containing vacuole (BCV) avoids fusion with lysosomes, prevents the deposition of complement at the bacterial surface and forms stable large clusters with MHC-II named macrodomians in the cell surface, interfering with MHC-II presentation of peptides to specific CD4+ T cells. Woah.
3. Listeria - It gets internalized in a vacuole and then runs away.

The pore-forming protein listeriolysin O mediates escape from host vacuoles. Once in the cytosol, the L. monocytogenes mediates efficient actin-based motility, thereby propelling the bacteria into neighboring cells. The cytosol is a favorable environment for listeria’s growth.
4. Mycobacterium leprae - Cholesterol and TACO!

Mycobacterium leprae is able to induce lipid droplet formation in infected macrophages. Cholesterol mediates the recruitment of TACO from the plasma membrane to the phagosome. TACO, also termed as coronin-1A (CORO1A), is a coat protein that prevents phagosome-lysosome fusion and thus degradation of mycobacteria in lysosomes. The entering of mycobacteria at cholesterol-rich domains of the plasma membrane and their subsequent uptake in TACO-coated phagosomes promotes intracellular survival.
5. Coxiella brunetti - The indestrucible

This hardy, obligate intracellular pathogen has evolved to not only survive, but to thrive, in the harshest of intracellular compartments: the phagolysosome. Following internalization, the nascent Coxiella phagosome ultimately develops into a large and spacious parasitophorous vacuole (PV) that acquires lysosomal characteristics such as acidic pH, acid hydrolases and cationic peptides, defences designed to rid the host of intruders.
6. Salmonella - TTSS

Salmonella have a specialized secretion system, termed the type III secretion system (TTSS), as well as proteins secreted by this system, are encoded in Salmonella pathogenicity island 1 (SPI1). TTSS are used by bacterial pathogens to inhibit their phagocytosis, induce eukaryotic cell death, and alter the host cell cytoskeleton. Salmonella species have at least one other TTSS encoded on SPI2 that appears to be involved in intracellular survival.
7. Human Immunodeficiency Virus - Tries to not attract attention

After infecting cells, HIV survives. Ever wondered why? It’s because the HIV protein, Nef plays a role in downregulating the expression of various proteins needed for recognition by potentially dangerous CD8 T cells. Nef lowers the surface expression of CD4, and several haplotypes of MHC-I by redirecting their transport from the trans-Golgi network. Another gene, Tat, appears to upregulate the expression of Bcl-2 during the early phase of cellular infection, increasing the likelihood that it will receive survival signals.
Many viruses can survive intracellularly, but I’ve included specifically HIV in this list because it survives in immune cells and it is an important virus to know.
For appropriate sources and references, click here.










Frankenstein’s…bacteriophages?
A group of scientists have just published their work in CellPress on synthesising novel phages using common viral structures (Figure 1), to target a range of bacterial hosts. Anyone who frequently reads my posts will be fully aware of what bacteriophages are, however put simply phages are viruses that target bacteria (Figure 1). Currently a lot of research is going into these little guys to utilise them for both therapeutic and diagnostic purposes.

Figure 1. Image depicts a transmission electron micrograph of a phage (left) and a diagrammatic representation of the phage on the (right) showing all the major components that make up the protein coat.
Phages have an elaborate protein coat that surrounds their DNA cargo (contained within the head, Figure 1). They scan their surrounding environment for potential ‘prey’ by using their tail fibres to interact with their cognate receptors. The diversity of receptors is immense and many still go unidentified even for well characterised phages. Once a target is found, the baseplate then irreversibly attaches to the cell wall of the bacteria and the DNA can be transferred into the cytoplasm of the cell.
Unlike current therapeutics, the sheer diversity between phages means that they are highly specific with limited host ranges to the species or even strain level. As a result, for phage therapy to be effective, a cocktail of phages needs to be employed to target a wide range of potential bacterial hosts. To improve the host ranges of phage groups such as this one are trying to synthesis novel structures from existing phages - much like how Frankenstein stitched together a new human body from pre-existing parts (Figure 2).

Figure 2. Simplified image showing how bacterial components can be shuffled between genomes of related phages with different host ranges.
Innovatively, this group used a well characterised yeast-based (Saccharomyces cerevisiae) platform for capturing phage genomes to allow their genetic manipulation (Figure 3). Phage genomes could then be inserted into a yeast artificial chromosome (YAC) then manipulated. Yeast are fairly easy to genetically modify by homologous-recombingation, making this system far easier to employ than other methods. The YAC was then recovered and transformed into the normal host bacteria, allowing the generation of new phage particles. Unlike other methods phage generated via this method were relatively easy to reboot back into active viral particles.

Figure 3. Illustration of the genetic manipulation of phage DNA to generate hybrid phages with altered host ranges.
The group managed to show that by swapping modular components such as tail fibres between phages, new host specificity could be generated. Thus illustrating that gene swapping can overcome strain or species barriers if the need arises. This work will hopefully lead way to further improve phage therapy and decrease the persistent need for the identification of novel phages. Further work needs to be done on increasing the scope of their work, but they have created a framework that will hopefully be able to reboot more synthetic phages.
“Our results show that common phage scaffolds can be re-targeted against new bacterial hosts by engineering single or multiple tail components. This capability enables the the construction of defined phage cocktails that only differ in their host range determinants and can be used to edit the compositions of microbial consortia and/or treat bacterial infections.” - Ando et al, 2015
Sources:
Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing, 2015. Ando, H., Lemire. S., Pires. D.P., Lu. T.K., Cell Systems , Volume 1 , Issue 3 , 187 - 196

JUST THE FAQS: Top Scientists Around the Nation Call for a Unified Microbiome Initiative
Writing in the journal Science this week, a group of top microbiome researchers from around the country, including UC San Diego School of Medicine’s Rob Knight, PhD, and Pieter Dorrestein, PhD, lay out a proposal for what they call a Unified Microbiome Initiative. This latest post in the JUST THE FAQS series takes a look at what that means and why it’s important.
What is the Unified Microbiome Initiative?
The proposed Unified Microbiome Initiative is a national effort to fund, coordinate and accelerate microbiome research — the study of the microbial communities (bacteria, viruses, etc.) that live in, on and around us.
Why do we need this?
According to the authors of this proposal, the microbiome field currently lacks many of the technologies and resources required to take the research from where it is today (i.e., mostly describing what microbes are living where) to the next level — understanding exactly how microbes influence our health and how we might be able to manipulate them to our benefit.
“… such knowledge could transform our understanding of the world and launch innovations in agriculture, energy, health, the environment and more,” the authors write in the proposal.
This ambitious undertaking cannot be accomplished by individual laboratories working in isolation. Developing these tools requires new collaborations between physical, life, and biomedical sciences, engineering and many other disciplines. Advancing this nascent field also depends on attracting, training and supporting multidisciplinary networks of scientists and engineers, all things that could be accomplished through a concerted national effort, Knight, Dorrestein and their co-authors say.
Wait, what is the microbiome again?
Your body contains as many as 10 times more microbial cells than human cells. Even crazier, that adds up to two to 20 million microbial genes, compared to your 20,000 or so human genes. By that measure, you could say you’re actually only about 10 percent human.
Researchers like Knight, Dorrestein and their teams are now mapping the other 90 percent — the collections of microbial genes (“microbiomes”) found in our guts, mouths, skin, and many other locations on and around the body. They’re finding that the makeup of our gut microbiomes is associated with a rapidly growing list of diseases and conditions: food allergies, obesity, inflammatory bowel disease and colon cancer, rheumatoid arthritis, atherosclerosis, asthma, perhaps even anxiety, depression and autism. But researchers don’t yet know why these associations occur, whether they are cause or effect, and whether or not we could use gut microbiome readouts for medical predictions, diagnoses or treatments.
And that’s just the gut microbiome. Imagine what researchers will find when they dive as deeply into the microbiomes of the rest of our bodies, as well as our homes, the ocean and other environments. That’s why Knight, Dorrestein and many other experts are today calling for a collaborative, well-supported United Microbiome Initiative.
To learn more about Knight’s and Dorrestein’s microbiome work, check out: Antibiotic Resistance in Microbiomes, Despite Isolation from Western Civilization
3D Human Skin Maps Aid Study of Relationships Between Molecules, Microbes and Environment
Pictured: Staphylococcus aureus bacteria. Source: NIAID


Bacterias & Tx
Hi guys, just wanted to let you know that I did this really fast, so if there’s something wrong let me know! thanks :)